Exploratory analysis
Model-based Geostatistics for Global Public Health
Overview of topics
- How to explore relationships with count data
- How to use a non-spatial model for mapping
- How to handle spatial data in R
Example: Riverblindness in Liberia
- How to explore relationships with count data (binary and aggregated counts)?
- How to model non-linear relationships?
- How to assess residual spatial correlation?
Overdispersion in Count Data
- What is overdispersion?
- Occurs when the variance of count data exceeds the mean.
- Violates the Poisson assumption:
\[ \text{Var}(Y) = \mathbb{E}(Y). \]
- Why does it matter?
- Standard models (e.g., Poisson regression) underestimate uncertainty.
- Leads to overly optimistic confidence intervals and p-values.
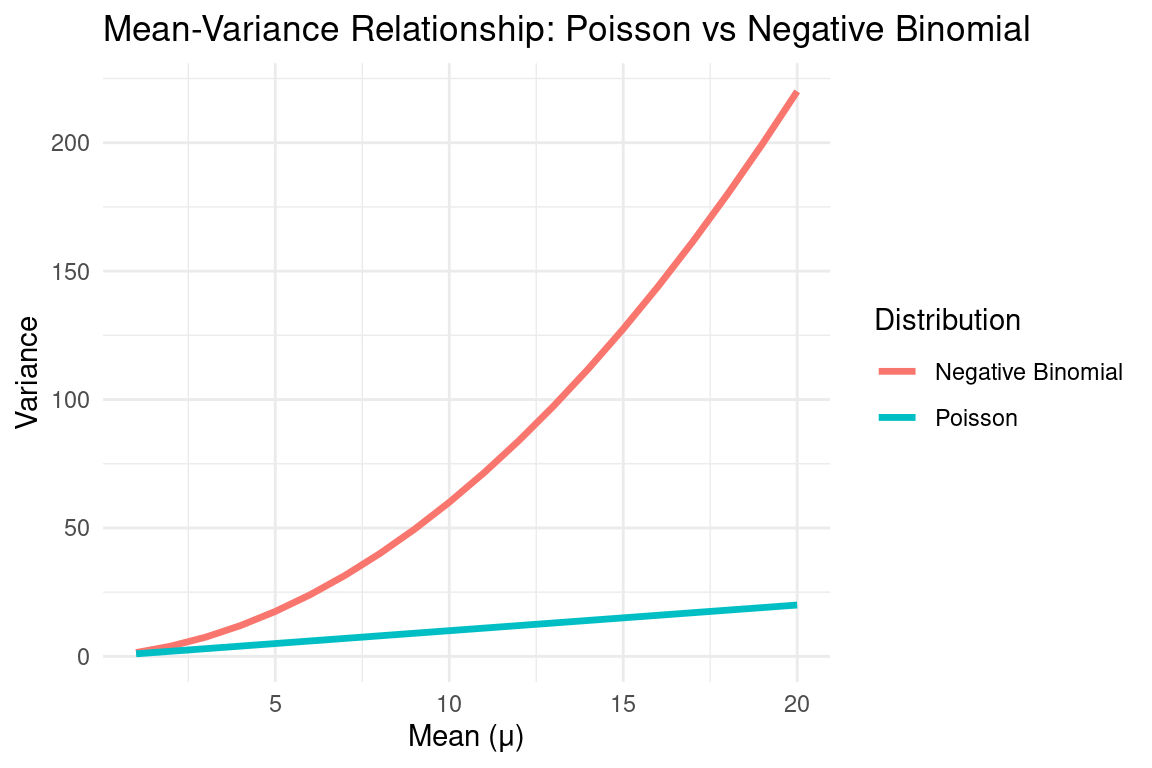
Example: Negative Binomial Distribution
A common solution for overdispersed count data.
Extends the Poisson distribution by introducing a dispersion parameter \(\alpha\):
\[ \mathbb{E}(Y) = \mu, \quad \text{Var}(Y) = \mu + \alpha \mu^2. \]When \(\alpha = 0\), it reduces to Poisson.
Random Effects Interpretation:
- The Negative Binomial can be interpreted as a Gamma-Poisson mixture.
- The Poisson mean \(\mu\) is drawn from a Gamma distribution, introducing extra variability.
Random Effects Models for Overdispersion
- Overdispersion often arises due to unobserved heterogeneity.
- A solution is to introduce random effects that account for latent variability.
. . .
- Random Intercept Model:
\[ Y_{ij} \mid Z_i \sim \text{Binomial}(p_{ij}), \quad \log\left\{\frac{p_{ij}}{1-p_{ij}}\right\} = \beta_0 + d_{ij} \beta_1 + Z_i. \]
. . .
- \(Z_i \sim \mathcal{N}(0, \sigma^2)\) captures between-group variability.
. . .
- Leads to extra variability in counts, addressing overdispersion.
. . .
- Connection to Negative Binomial:
- If \(Z_i\) follows a Gamma distribution instead of Normal, the model is equivalent to a Negative Binomial.
A class of generalized linear models
Assumptions:
. . .
- \(Z_{i}\) are i.i.d. random variables;
. . .
- \(Y_{i} \mid Z_i \sim f(\cdot)\) belongs to the exponential family;
. . .
- \(E[Y_{i}\mid Z_i] = m_i \mu_{i}\) and \(\text{Var}[Y_{i} \mid Z_i] = m_i V(\mu_{i})\);
. . .
- \(g(\mu_{i}) = \eta_i = d_i^\top \beta + Z_i\);
. . .
- \(Y_i \mid Z_i\) are mutually independent for \(i=1,\dots,n\).
Parameter Estimation
Let the \(Z_i\) be i.i.d. Gaussian distributions with mean zero and variance \(\sigma^2\).
Likelihood function
The vector of unkown parameters is \(\theta=(\beta, \sigma^2)\)
\[ L(\theta) = \prod_{i=1}^n \int_{-\infty}^{+\infty} [Z_i] [Y_i \mid Z_i] \: dY_i \]
. . .
- Estimation Method
Maximize the likelihood using the Laplace approximation (glmer in the lme4 package).
. . .
Hypothesis Testing
- Obtain \(\hat{\theta}\) (MLE).
- Obtain \(\hat{\theta}_{0}\), the MLE constrained by fixing \(p\) values of \(\beta\) to 0.
- Compute the log-likelihood ratio:
\[ D = 2(\log L(\hat{\theta}) - \log L(\hat{\theta}_0)) \sim \chi^2_{p} \]
- P-value: \(P(D > D_{obs} \mid H_0)\)
. . .
Which residuals should we use to assess spatial correlation?
Pearson Residuals: \[\frac{y_i - \hat{y}_i}{\sqrt{\text{Var}(\hat{y}_i)}}\]
Deviance Residuals: \[\text{sign}(y_i - \hat{y}_i) \times \sqrt{d_i}\] where \(d_i\) is the deviance contribution of observation \(i\).
Random Effects Residuals: \(\hat{Z}_i\), i.e. the estimated random effect for each location/household/cluster/village.
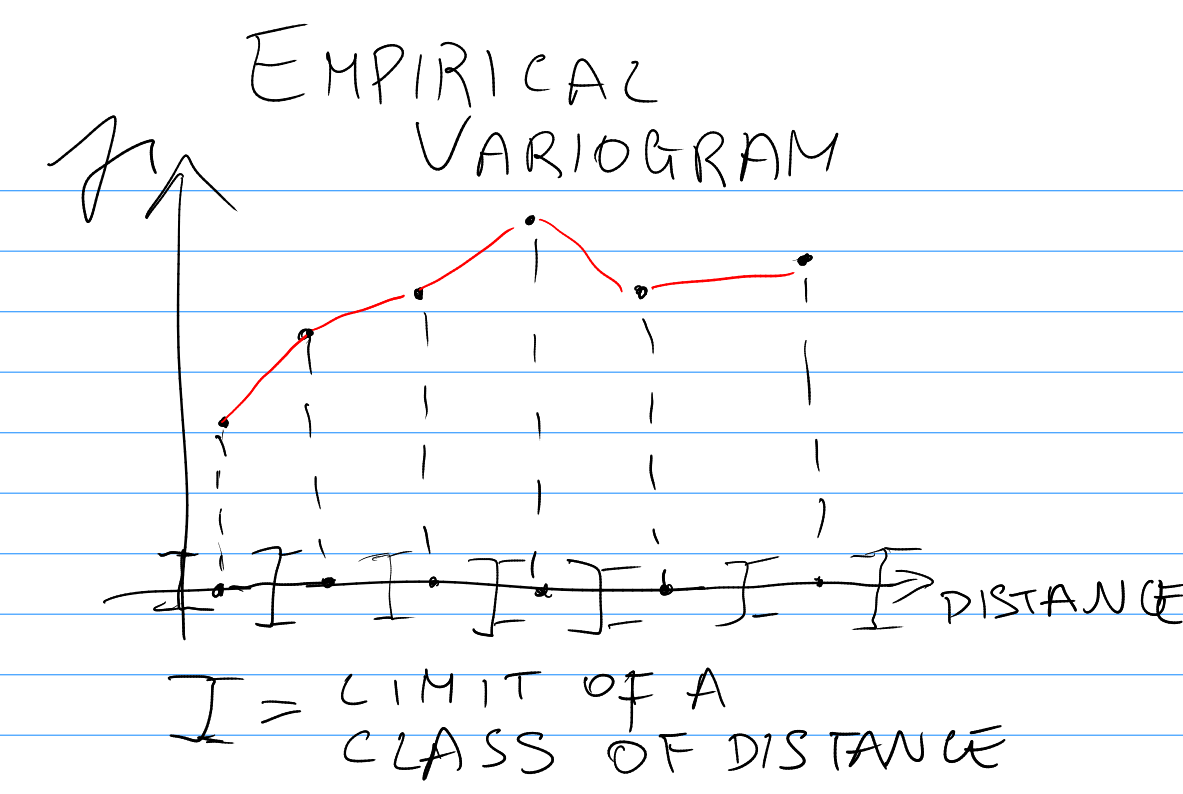
The Empirical Variogram using Random Effects Residuals
- The empirical variogram is defined as: \[
\gamma(D_{[a,b]}) = \frac{1}{2|D_{[a,b]}|} \sum_{(i,j) \in D_{[a,b]}} (\hat{Z}_i - \hat{Z}_j)^2
\] where:
- \(D_{[a,b]}\) is a class of distance with lower limit \(a\) and upper limit \(b\);
- \(|D_{[a,b]}|\) is the number of pairs of observations whose locations ar at a distance between \(a\) and \(b\).
. . .
. . .
Coordinate Reference Systems (CRS)
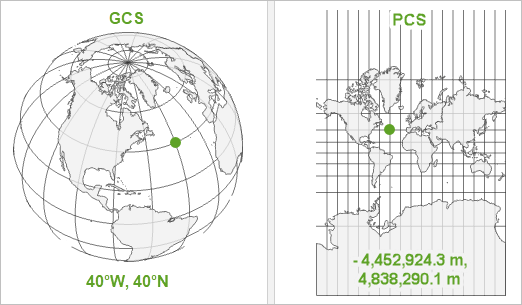
- A CRS tells us how the coordinates in our data match real-world locations on the Earth.
- Two main types:
- Geographic CRS (Latitude/Longitude, e.g., WGS84)
- Projected CRS (e.g., UTM, which uses meters for measurements)
- Geographic CRS (Latitude/Longitude, e.g., WGS84)
Computing Distance
Distances in Geographic CRS (Lat/Lon) require geodesic calculations (e.g., Haversine formula).
Projecting to UTM allows for Euclidean distance calculations in meters.
Conversion to UTM in R:
library(sf) points <- st_as_sf(data.frame(lon = c(-0.1, -0.2), lat = c(51.5, 51.6)), coords = c("lon", "lat"), crs = 4326) points_utm <- st_transform(points, crs = 32630)
Spatial Data Formats
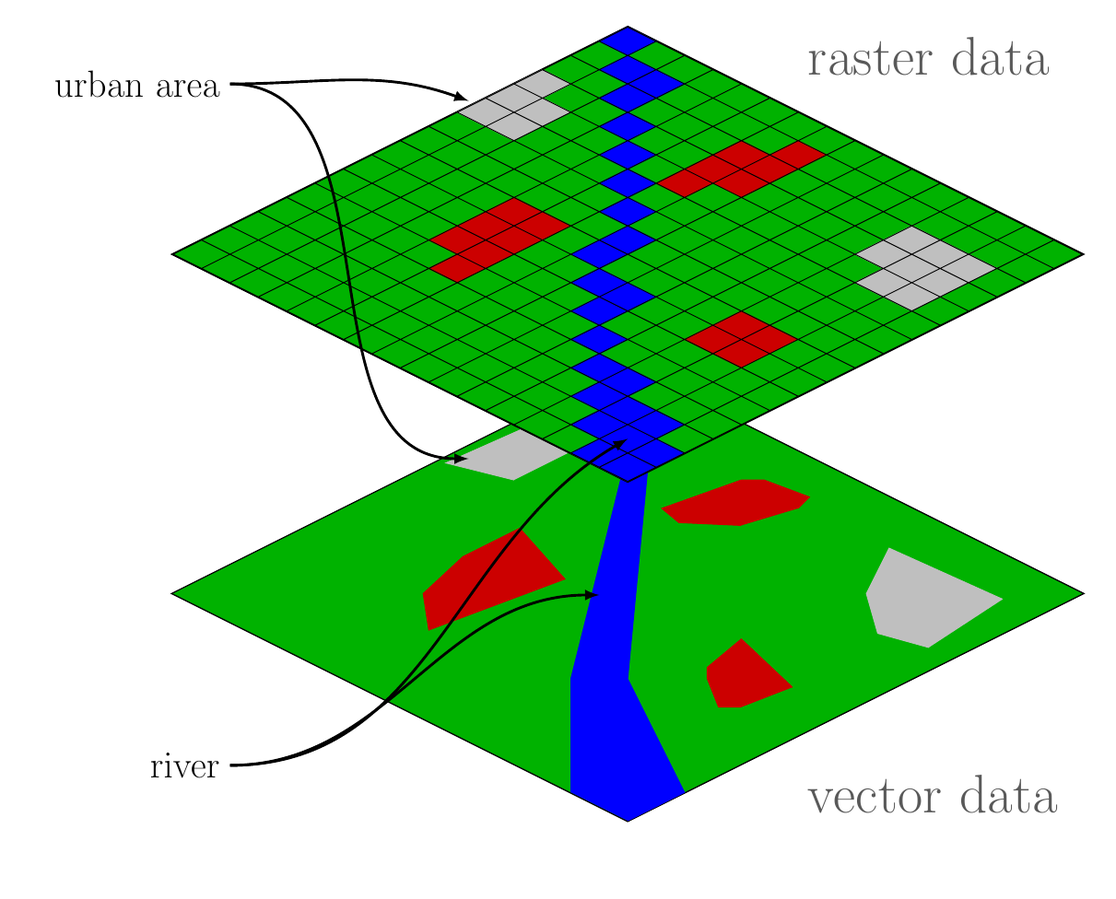
- Shapefiles:
- Vector data format for storing geometries (points, lines, polygons).
- Commonly used in GIS applications.
- Raster Files:
Grid-based data (e.g., satellite images, elevation models).
Each cell has a value representing an attribute (e.g., temperature, elevation).
